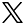Our laboratory aims to understand how neurons differentiate and assemble into circuits.


Adrian Moore, Ph.D.
Team Director, Neurodiversity
adrian.moore@riken.jp
Research Overview
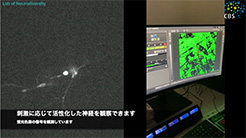
Neurons form among the most complex and diverse structures of any cell type. Our laboratory aims to understand how neurons differentiate and assemble into circuits. While we can now create a list of neuron types and a connectivity map for the brain, we still do not understand the regulatory mechanisms by which neuron differentiation is genetically encoded, nor how morphogenetic and cell biological events are controlled to create complex highly branched dendritic (receiving) and axonal (sending) structures that create the circuits in the brain. Understanding these questions is vital because imbalances among neuron types and disrupted dendrite formation lead to neurodevelopmental disorders. Unfortunately, these disorders are common, with approximately 2% of children developing one or both intellectual disability and autism spectrum disorder. Moreover, if impaired, failed neuron differentiation can also prime cells for age-related neurodegeneration.
To address these questions, my group has developed an interdisciplinary approach that combines genetics, genomics, biochemistry, in vivo imaging of neuron differentiation, computational image analysis, and quantitative cell biology. In vivo, using Drosophila, we visualize, manipulate, and measure the structure of neurons as they differentiate their complex branched connections in their natural environment. Critically, because there is a high degree of conservation at the molecular level between Drosophila and human neurogenesis, we can use our mechanistic insight gained from these powerful in vivo tools in Drosophila to guide complementary studies using human neuronal stem cell and neuron culture systems.
Main Research Fields
Biological Sciences
Related Research Fields
Interdisciplinary Science & Engineering / Biological Sciences / Biology / Medicine, Dentistry & Pharmacy / Neuroscience-general-related / Cell biology-related / Developmental biology-related
Keywords
- neuron differentiation
- dendrite and axon
- neural stem cells
- microtubules
- in vivo cell imaging
Selected Publications
- Suzuki T., Tatsukawa T., Sudo G., Delandre C., Pai YJ., Miyamoto H., Raveau M., Shimohata A., Ohmori I., Hamano S., Haginoya K., Uematsu M., Takahashi Y., Morimoto M., Fujimoto S., Osaka H., Oguni H., Osawa M., Ishii A., Hirose S., Kaneko S., Inoue Y., Moore AW., Yamakawa K.
"CUX2 deficiency causes facilitation of excitatory synaptic transmission onto hippocampus and increased seizure susceptibility to kainite"
Scientific Reports 12, e6505. (2022). - Pai YJ., Moore AW.
"Transcription factor encoding of neuron subtype: strategies that specify arbor pattern"
Curr. Opin. Neurobiol. 69, pp149-158. (2021). - Yoong LF., Lim HK., Tran H., Lackner S., Zheng Z., Hong P., Moore AW.
"Atypical myosin tunes dendrite arbor subdivision"
Neuron 106, pp452-467. (2020). - Rives-Quinto N., Komori N., Ostgaard CM., Janssens DH., Kondo S., Dai Q., Moore AW., Lee CY.
"Sequential activation of transcriptional repressors promotes progenitor commitment by silencing stem cell identity genes"
Elife 9, e56187. (2020). - Liang X., Kokes M., Fetter RD., Sallee MD., Moore AW., Feldman JL., Shen K.
"Growth cone-localized microtubule organizing center establishes microtubule orientation in dendrites"
Elife 9, e56547. (2020). - Wilkes OR., Moore AW.
"Distinct Microtubule Organizing Center Mechanisms Combine to Generate Neuron Polarity and Arbor Complexity"
Front. Cell. Neurosci. 14, e398. (2020). - Urun FR., Moore AW.
"Visualizing Cell Cycle Phase Organization and Control During Neural Lineage Elaboration"
Cells 9, e2112. (2020). - Chen YC., Auer-Grumbach M., Matsukawa S., Zitzelsberger M., Themistocleous AC., Strom TM., Samara C., Moore AW., Cho LT., Young GT., Weiss C., Schabhüttl M., Stucka R., Schmid AB., Parman Y., Graul-Neumann L., Heinritz W., Passarge E., Watson RM., Hertz JM., Moog U., Baumgartner M., Valente EM., Pereira D., Restrepo CM., Katona I., Dusl M., Stendel C., Wieland T., Stafford F., Reimann F., von Au K., Finke C., Willems PJ., Nahorski MS., Shaikh SS., Carvalho OP., Nicholas AK., Karbani G., McAleer MA., Cilio MR., McHugh JC., Murphy SM., Irvine AD., Jensen UB., Windhager R., Weis J., Bergmann C., Rautenstrauss B., Baets J., De Jonghe P., Reilly MM., Kropatsch R., Kurth I., Chrast R., Michiue T., Bennett DL., Woods CG., Senderek J.
"Transcriptional regulator PRDM12 is essential for human pain perception"
Nat. Genet. 7, pp803-8. (2015) - Yalgin C., Ebrahimi S., Delandre C., Yoong LF., Akimoto S., Tran H., Amikura R., Spokony R., Torben-Nielsen B., White KP., Moore AW.
"Centrosomin represses dendrite branching by orienting microtubule nucleation"
Nat. Neurosci. 18, pp1437-45. (2015). - Endo K., Karim MR., Taniguchi H., Krejci A., Kinameri E., Siebert M., Ito K., Bray S., Moore AW.
"Chromatin modification of Notch targets in olfactory receptor neuron diversification"
Nat. Neurosci. 15, pp224-33. (2012).
Lab Members
Principal investigator
- Adrian Walton Moore
- Team Director
Core members
- Fangke Xu
- Postdoctoral Researcher
- Minami Kimura
- Research Part-time Worker I